Data Wrangling with Pandas II
Contents
Data Wrangling with Pandas II#

Outline#
In this class we will talk more about:
“Tips and tricks” for working with
pandasHandling Missing Data
Combining Datasets: Concat and Append
Visualizing Data using
seabornandpandasAdditional Notes you may found helpful
Working with Pandas to load data#
import pandas as pd
Always use RELATIVE paths!!#
pwd
'/home/runner/work/data301_course/data301_course/notes/week05/Class5B'
!ls data/
chord-fingers1.csv state-abbrevs.csv state-population.csv
chord-fingers2.csv state-areas.csv
What if your data is not UTF-8 encoded?#
(Hint: You will get a UnicodeDecodeError)
url = "http://nfdp.ccfm.org/download/data/csv/NFD%20-%20Area%20burned%20by%20cause%20class%20-%20EN%20FR.csv"
dataset = pd.read_csv(url)
---------------------------------------------------------------------------
UnicodeDecodeError Traceback (most recent call last)
Cell In[4], line 3
1 url = "http://nfdp.ccfm.org/download/data/csv/NFD%20-%20Area%20burned%20by%20cause%20class%20-%20EN%20FR.csv"
----> 3 dataset = pd.read_csv(url)
File /opt/hostedtoolcache/Python/3.10.9/x64/lib/python3.10/site-packages/pandas/util/_decorators.py:211, in deprecate_kwarg.<locals>._deprecate_kwarg.<locals>.wrapper(*args, **kwargs)
209 else:
210 kwargs[new_arg_name] = new_arg_value
--> 211 return func(*args, **kwargs)
File /opt/hostedtoolcache/Python/3.10.9/x64/lib/python3.10/site-packages/pandas/util/_decorators.py:331, in deprecate_nonkeyword_arguments.<locals>.decorate.<locals>.wrapper(*args, **kwargs)
325 if len(args) > num_allow_args:
326 warnings.warn(
327 msg.format(arguments=_format_argument_list(allow_args)),
328 FutureWarning,
329 stacklevel=find_stack_level(),
330 )
--> 331 return func(*args, **kwargs)
File /opt/hostedtoolcache/Python/3.10.9/x64/lib/python3.10/site-packages/pandas/io/parsers/readers.py:950, in read_csv(filepath_or_buffer, sep, delimiter, header, names, index_col, usecols, squeeze, prefix, mangle_dupe_cols, dtype, engine, converters, true_values, false_values, skipinitialspace, skiprows, skipfooter, nrows, na_values, keep_default_na, na_filter, verbose, skip_blank_lines, parse_dates, infer_datetime_format, keep_date_col, date_parser, dayfirst, cache_dates, iterator, chunksize, compression, thousands, decimal, lineterminator, quotechar, quoting, doublequote, escapechar, comment, encoding, encoding_errors, dialect, error_bad_lines, warn_bad_lines, on_bad_lines, delim_whitespace, low_memory, memory_map, float_precision, storage_options)
935 kwds_defaults = _refine_defaults_read(
936 dialect,
937 delimiter,
(...)
946 defaults={"delimiter": ","},
947 )
948 kwds.update(kwds_defaults)
--> 950 return _read(filepath_or_buffer, kwds)
File /opt/hostedtoolcache/Python/3.10.9/x64/lib/python3.10/site-packages/pandas/io/parsers/readers.py:605, in _read(filepath_or_buffer, kwds)
602 _validate_names(kwds.get("names", None))
604 # Create the parser.
--> 605 parser = TextFileReader(filepath_or_buffer, **kwds)
607 if chunksize or iterator:
608 return parser
File /opt/hostedtoolcache/Python/3.10.9/x64/lib/python3.10/site-packages/pandas/io/parsers/readers.py:1442, in TextFileReader.__init__(self, f, engine, **kwds)
1439 self.options["has_index_names"] = kwds["has_index_names"]
1441 self.handles: IOHandles | None = None
-> 1442 self._engine = self._make_engine(f, self.engine)
File /opt/hostedtoolcache/Python/3.10.9/x64/lib/python3.10/site-packages/pandas/io/parsers/readers.py:1753, in TextFileReader._make_engine(self, f, engine)
1750 raise ValueError(msg)
1752 try:
-> 1753 return mapping[engine](f, **self.options)
1754 except Exception:
1755 if self.handles is not None:
File /opt/hostedtoolcache/Python/3.10.9/x64/lib/python3.10/site-packages/pandas/io/parsers/c_parser_wrapper.py:79, in CParserWrapper.__init__(self, src, **kwds)
76 kwds.pop(key, None)
78 kwds["dtype"] = ensure_dtype_objs(kwds.get("dtype", None))
---> 79 self._reader = parsers.TextReader(src, **kwds)
81 self.unnamed_cols = self._reader.unnamed_cols
83 # error: Cannot determine type of 'names'
File /opt/hostedtoolcache/Python/3.10.9/x64/lib/python3.10/site-packages/pandas/_libs/parsers.pyx:547, in pandas._libs.parsers.TextReader.__cinit__()
File /opt/hostedtoolcache/Python/3.10.9/x64/lib/python3.10/site-packages/pandas/_libs/parsers.pyx:636, in pandas._libs.parsers.TextReader._get_header()
File /opt/hostedtoolcache/Python/3.10.9/x64/lib/python3.10/site-packages/pandas/_libs/parsers.pyx:852, in pandas._libs.parsers.TextReader._tokenize_rows()
File /opt/hostedtoolcache/Python/3.10.9/x64/lib/python3.10/site-packages/pandas/_libs/parsers.pyx:1965, in pandas._libs.parsers.raise_parser_error()
UnicodeDecodeError: 'utf-8' codec can't decode byte 0xe9 in position 8: invalid continuation byte
Demo#
Now what!?
Now we google it…
And according to this stack overflow page: we should try:
pd.read_csv(url, encoding='latin')
So let’s give it a shot:
pd.read_csv(url, encoding="latin")
# Works!!
| Year | Année | ISO | Jurisdiction | Juridiction | Cause | Origine | Area (hectares) | Data Qualifier | Superficie (en hectare) | Qualificatifs de données | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1990 | 1990 | AB | Alberta | Alberta | Human activity | Activités humaines | 2393.8 | a | 2393.8 | a |
| 1 | 1990 | 1990 | AB | Alberta | Alberta | Lightning | Foudre | 55482.6 | a | 55482.6 | a |
| 2 | 1990 | 1990 | AB | Alberta | Alberta | Unspecified | Indéterminée | 1008.8 | a | 1008.8 | a |
| 3 | 1990 | 1990 | BC | British Columbia | Colombie-Britannique | Human activity | Activités humaines | 40278.3 | a | 40278.3 | a |
| 4 | 1990 | 1990 | BC | British Columbia | Colombie-Britannique | Lightning | Foudre | 35503.5 | a | 35503.5 | a |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1054 | 2021 | 2021 | PC | Parks Canada | Parcs Canada | Unspecified | Indéterminée | 42538.0 | e | 42538.0 | e |
| 1055 | 2021 | 2021 | PE | Prince Edward Island | Île-du-Prince-Édouard | Unspecified | Indéterminée | 0.1 | e | 0.1 | e |
| 1056 | 2021 | 2021 | QC | Quebec | Québec | Unspecified | Indéterminée | 49748.0 | e | 49748.0 | e |
| 1057 | 2021 | 2021 | SK | Saskatchewan | Saskatchewan | Unspecified | Indéterminée | 956084.0 | e | 956084.0 | e |
| 1058 | 2021 | 2021 | YT | Yukon | Yukon | Unspecified | Indéterminée | 118126.0 | e | 118126.0 | e |
1059 rows × 11 columns
What if your data is not comma-separated?#
chord1 = pd.read_csv("data/chord-fingers1.csv")
chord1.head()
---------------------------------------------------------------------------
ParserError Traceback (most recent call last)
Input In [7], in <cell line: 1>()
----> 1 chord1 = pd.read_csv("data/chord-fingers1.csv")
2 chord1.head()
File ~/.pyenv/versions/3.10.2/lib/python3.10/site-packages/pandas/util/_decorators.py:311, in deprecate_nonkeyword_arguments.<locals>.decorate.<locals>.wrapper(*args, **kwargs)
305 if len(args) > num_allow_args:
306 warnings.warn(
307 msg.format(arguments=arguments),
308 FutureWarning,
309 stacklevel=stacklevel,
310 )
--> 311 return func(*args, **kwargs)
File ~/.pyenv/versions/3.10.2/lib/python3.10/site-packages/pandas/io/parsers/readers.py:680, in read_csv(filepath_or_buffer, sep, delimiter, header, names, index_col, usecols, squeeze, prefix, mangle_dupe_cols, dtype, engine, converters, true_values, false_values, skipinitialspace, skiprows, skipfooter, nrows, na_values, keep_default_na, na_filter, verbose, skip_blank_lines, parse_dates, infer_datetime_format, keep_date_col, date_parser, dayfirst, cache_dates, iterator, chunksize, compression, thousands, decimal, lineterminator, quotechar, quoting, doublequote, escapechar, comment, encoding, encoding_errors, dialect, error_bad_lines, warn_bad_lines, on_bad_lines, delim_whitespace, low_memory, memory_map, float_precision, storage_options)
665 kwds_defaults = _refine_defaults_read(
666 dialect,
667 delimiter,
(...)
676 defaults={"delimiter": ","},
677 )
678 kwds.update(kwds_defaults)
--> 680 return _read(filepath_or_buffer, kwds)
File ~/.pyenv/versions/3.10.2/lib/python3.10/site-packages/pandas/io/parsers/readers.py:581, in _read(filepath_or_buffer, kwds)
578 return parser
580 with parser:
--> 581 return parser.read(nrows)
File ~/.pyenv/versions/3.10.2/lib/python3.10/site-packages/pandas/io/parsers/readers.py:1254, in TextFileReader.read(self, nrows)
1252 nrows = validate_integer("nrows", nrows)
1253 try:
-> 1254 index, columns, col_dict = self._engine.read(nrows)
1255 except Exception:
1256 self.close()
File ~/.pyenv/versions/3.10.2/lib/python3.10/site-packages/pandas/io/parsers/c_parser_wrapper.py:225, in CParserWrapper.read(self, nrows)
223 try:
224 if self.low_memory:
--> 225 chunks = self._reader.read_low_memory(nrows)
226 # destructive to chunks
227 data = _concatenate_chunks(chunks)
File pandas/_libs/parsers.pyx:805, in pandas._libs.parsers.TextReader.read_low_memory()
File pandas/_libs/parsers.pyx:861, in pandas._libs.parsers.TextReader._read_rows()
File pandas/_libs/parsers.pyx:847, in pandas._libs.parsers.TextReader._tokenize_rows()
File pandas/_libs/parsers.pyx:1960, in pandas._libs.parsers.raise_parser_error()
ParserError: Error tokenizing data. C error: Expected 10 fields in line 7, saw 11
Demo#
Now what!?
Now we google the error message…
And according to this stack overflow page: we should try:
pd.read_csv('data/chord-fingers1.csv', sep=';')
So let’s give it a shot:
chord1 = pd.read_csv("data/chord-fingers1.csv", sep=";")
chord1.head()
| CHORD_ROOT | CHORD_TYPE | CHORD_STRUCTURE | FINGER_POSITIONS | NOTE_NAMES | |
|---|---|---|---|---|---|
| 0 | A# | 13 | 1;3;5;b7;9;11;13 | x,1,0,2,3,4 | A#,C##,G#,B#,F## |
| 1 | A# | 13 | 1;3;5;b7;9;11;13 | 4,x,3,2,1,1 | A#,G#,B#,C##,F## |
| 2 | A# | 13 | 1;3;5;b7;9;11;13 | 1,x,1,2,3,4 | A#,G#,C##,F##,B# |
| 3 | A# | 7(#9) | 1;3;5;b7;#9 | x,1,0,2,4,3 | A#,C##,G#,B##,E# |
| 4 | A# | 7(#9) | 1;3;5;b7;#9 | 2,1,3,3,3,x | A#,C##,G#,B##,E# |
# Let's load in the second part of the dataset in the same way:
chord2 = pd.read_csv("data/chord-fingers2.csv", sep=";")
chord2.head()
| CHORD_ROOT | CHORD_TYPE | CHORD_STRUCTURE | FINGER_POSITIONS | NOTE_NAMES | |
|---|---|---|---|---|---|
| 0 | Ab | 6 | 1;3;5;6 | x,4,2,3,1,x | Ab,C,F,Ab |
| 1 | Ab | 6 | 1;3;5;6 | x,x,3,2,4,1 | Ab,C,F,Ab |
| 2 | Ab | 6 | 1;3;5;6 | 2,x,1,4,x,x | Ab,F,C |
| 3 | Ab | 6 | 1;3;5;6 | 1,x,x,3,4,x | Ab,C,F |
| 4 | Ab | 6 | 1;3;5;6 | x,x,1,3,1,4 | Ab,Eb,F,C |
len(chord1)
598
len(chord2)
2034
len(chord1) + len(chord2)
2632
Combining DataFrames together#
We saw that our dataset came in two parts (chord1 and chord2).
A common task in data analysis is to merge or combine datasets together. There are several combinations we can do.
Let’s look at just a basic one:
Using .concat()#
Documentation on the .concat() function

# chord1['New Column'] ='Firas'
chord1
| CHORD_ROOT | CHORD_TYPE | CHORD_STRUCTURE | FINGER_POSITIONS | NOTE_NAMES | |
|---|---|---|---|---|---|
| 0 | A# | 13 | 1;3;5;b7;9;11;13 | x,1,0,2,3,4 | A#,C##,G#,B#,F## |
| 1 | A# | 13 | 1;3;5;b7;9;11;13 | 4,x,3,2,1,1 | A#,G#,B#,C##,F## |
| 2 | A# | 13 | 1;3;5;b7;9;11;13 | 1,x,1,2,3,4 | A#,G#,C##,F##,B# |
| 3 | A# | 7(#9) | 1;3;5;b7;#9 | x,1,0,2,4,3 | A#,C##,G#,B##,E# |
| 4 | A# | 7(#9) | 1;3;5;b7;#9 | 2,1,3,3,3,x | A#,C##,G#,B##,E# |
| ... | ... | ... | ... | ... | ... |
| 593 | Ab | m7 | 1;b3;5;b7 | 1,3,1,1,1,x | Ab,Eb,Gb,Cb,Eb |
| 594 | Ab | dim | 1;b3;b5 | x,x,4,3,1,2 | Ab,Cb,Ebb,Ab |
| 595 | Ab | dim | 1;b3;b5 | x,1,2,4,3,x | Ab,Ebb,Ab,Cb |
| 596 | Ab | 6 | 1;3;5;6 | x,x,1,1,1,1 | Eb,Ab,C,F |
| 597 | Ab | 6 | 1;3;5;6 | 1,x,3,2,4,1 | Ab,Ab,C,F,Ab |
598 rows × 5 columns
pd.concat([chord1, chord2])
| CHORD_ROOT | CHORD_TYPE | CHORD_STRUCTURE | FINGER_POSITIONS | NOTE_NAMES | New Column | |
|---|---|---|---|---|---|---|
| 0 | A# | 13 | 1;3;5;b7;9;11;13 | x,1,0,2,3,4 | A#,C##,G#,B#,F## | Firas |
| 1 | A# | 13 | 1;3;5;b7;9;11;13 | 4,x,3,2,1,1 | A#,G#,B#,C##,F## | Firas |
| 2 | A# | 13 | 1;3;5;b7;9;11;13 | 1,x,1,2,3,4 | A#,G#,C##,F##,B# | Firas |
| 3 | A# | 7(#9) | 1;3;5;b7;#9 | x,1,0,2,4,3 | A#,C##,G#,B##,E# | Firas |
| 4 | A# | 7(#9) | 1;3;5;b7;#9 | 2,1,3,3,3,x | A#,C##,G#,B##,E# | Firas |
| ... | ... | ... | ... | ... | ... | ... |
| 2029 | G | sus4 | 1;4;5 | x,1,2,3,4,1 | G,D,G,C,D | NaN |
| 2030 | G | sus4 | 1;4;5 | x,x,3,4,1,1 | G,C,D,G | NaN |
| 2031 | G | 6/9 | 1;3;5;6;9 | 2,1,1,1,3,4 | G,B,E,A,D,G | NaN |
| 2032 | G | 6/9 | 1;3;5;6;9 | x,x,2,1,3,4 | G,B,E,A | NaN |
| 2033 | G | 6/9 | 1;3;5;6;9 | x,2,1,1,3,4 | G,B,E,A,D | NaN |
2632 rows × 6 columns
# You NEED to save the output of all pandas function, do NOT use the "inplace" argument, it will be deprecated soon
chords = pd.concat([chord1, chord2])
chords
| CHORD_ROOT | CHORD_TYPE | CHORD_STRUCTURE | FINGER_POSITIONS | NOTE_NAMES | |
|---|---|---|---|---|---|
| 0 | A# | 13 | 1;3;5;b7;9;11;13 | x,1,0,2,3,4 | A#,C##,G#,B#,F## |
| 1 | A# | 13 | 1;3;5;b7;9;11;13 | 4,x,3,2,1,1 | A#,G#,B#,C##,F## |
| 2 | A# | 13 | 1;3;5;b7;9;11;13 | 1,x,1,2,3,4 | A#,G#,C##,F##,B# |
| 3 | A# | 7(#9) | 1;3;5;b7;#9 | x,1,0,2,4,3 | A#,C##,G#,B##,E# |
| 4 | A# | 7(#9) | 1;3;5;b7;#9 | 2,1,3,3,3,x | A#,C##,G#,B##,E# |
| ... | ... | ... | ... | ... | ... |
| 2029 | G | sus4 | 1;4;5 | x,1,2,3,4,1 | G,D,G,C,D |
| 2030 | G | sus4 | 1;4;5 | x,x,3,4,1,1 | G,C,D,G |
| 2031 | G | 6/9 | 1;3;5;6;9 | 2,1,1,1,3,4 | G,B,E,A,D,G |
| 2032 | G | 6/9 | 1;3;5;6;9 | x,x,2,1,3,4 | G,B,E,A |
| 2033 | G | 6/9 | 1;3;5;6;9 | x,2,1,1,3,4 | G,B,E,A,D |
2632 rows × 5 columns
Using .rename()#
Documentation on the .rename() function:
# Capitals annoy me!
list(chords.columns)
['CHORD_ROOT',
'CHORD_TYPE',
'CHORD_STRUCTURE',
'FINGER_POSITIONS',
'NOTE_NAMES']
chords = chords.rename(
)
chords.head()
| Chord | Type | Structure | Finger | Notes | |
|---|---|---|---|---|---|
| 0 | A# | 13 | 1;3;5;b7;9;11;13 | x,1,0,2,3,4 | A#,C##,G#,B#,F## |
| 1 | A# | 13 | 1;3;5;b7;9;11;13 | 4,x,3,2,1,1 | A#,G#,B#,C##,F## |
| 2 | A# | 13 | 1;3;5;b7;9;11;13 | 1,x,1,2,3,4 | A#,G#,C##,F##,B# |
| 3 | A# | 7(#9) | 1;3;5;b7;#9 | x,1,0,2,4,3 | A#,C##,G#,B##,E# |
| 4 | A# | 7(#9) | 1;3;5;b7;#9 | 2,1,3,3,3,x | A#,C##,G#,B##,E# |
Using .replace()#
Documentation on the .replace() function:
chords_cleaned = chords.replace("A#", "A-sharp")
chords_cleaned
| Chord | Type | Structure | Finger | Notes | |
|---|---|---|---|---|---|
| 0 | A-sharp | 13 | 1;3;5;b7;9;11;13 | x,1,0,2,3,4 | A#,C##,G#,B#,F## |
| 1 | A-sharp | 13 | 1;3;5;b7;9;11;13 | 4,x,3,2,1,1 | A#,G#,B#,C##,F## |
| 2 | A-sharp | 13 | 1;3;5;b7;9;11;13 | 1,x,1,2,3,4 | A#,G#,C##,F##,B# |
| 3 | A-sharp | 7(#9) | 1;3;5;b7;#9 | x,1,0,2,4,3 | A#,C##,G#,B##,E# |
| 4 | A-sharp | 7(#9) | 1;3;5;b7;#9 | 2,1,3,3,3,x | A#,C##,G#,B##,E# |
| ... | ... | ... | ... | ... | ... |
| 2029 | G | sus4 | 1;4;5 | x,1,2,3,4,1 | G,D,G,C,D |
| 2030 | G | sus4 | 1;4;5 | x,x,3,4,1,1 | G,C,D,G |
| 2031 | G | 6/9 | 1;3;5;6;9 | 2,1,1,1,3,4 | G,B,E,A,D,G |
| 2032 | G | 6/9 | 1;3;5;6;9 | x,x,2,1,3,4 | G,B,E,A |
| 2033 | G | 6/9 | 1;3;5;6;9 | x,2,1,1,3,4 | G,B,E,A,D |
2632 rows × 5 columns
Using .drop()#
Documentation on the .drop() function:
chords_cleaned
| Chord | Type | Structure | Finger | Notes | |
|---|---|---|---|---|---|
| 0 | A-sharp | 13 | 1;3;5;b7;9;11;13 | x,1,0,2,3,4 | A#,C##,G#,B#,F## |
| 1 | A-sharp | 13 | 1;3;5;b7;9;11;13 | 4,x,3,2,1,1 | A#,G#,B#,C##,F## |
| 2 | A-sharp | 13 | 1;3;5;b7;9;11;13 | 1,x,1,2,3,4 | A#,G#,C##,F##,B# |
| 3 | A-sharp | 7(#9) | 1;3;5;b7;#9 | x,1,0,2,4,3 | A#,C##,G#,B##,E# |
| 4 | A-sharp | 7(#9) | 1;3;5;b7;#9 | 2,1,3,3,3,x | A#,C##,G#,B##,E# |
| ... | ... | ... | ... | ... | ... |
| 2029 | G | sus4 | 1;4;5 | x,1,2,3,4,1 | G,D,G,C,D |
| 2030 | G | sus4 | 1;4;5 | x,x,3,4,1,1 | G,C,D,G |
| 2031 | G | 6/9 | 1;3;5;6;9 | 2,1,1,1,3,4 | G,B,E,A,D,G |
| 2032 | G | 6/9 | 1;3;5;6;9 | x,x,2,1,3,4 | G,B,E,A |
| 2033 | G | 6/9 | 1;3;5;6;9 | x,2,1,1,3,4 | G,B,E,A,D |
2632 rows × 5 columns
chords_cleaned = chords_cleaned.drop(["Notes", "New Column"], axis="columns")
sorted(list(chords_cleaned.columns))
['Chord', 'Finger', 'Structure', 'Type']
chords = chords[sorted(list(chords.columns))]
chords
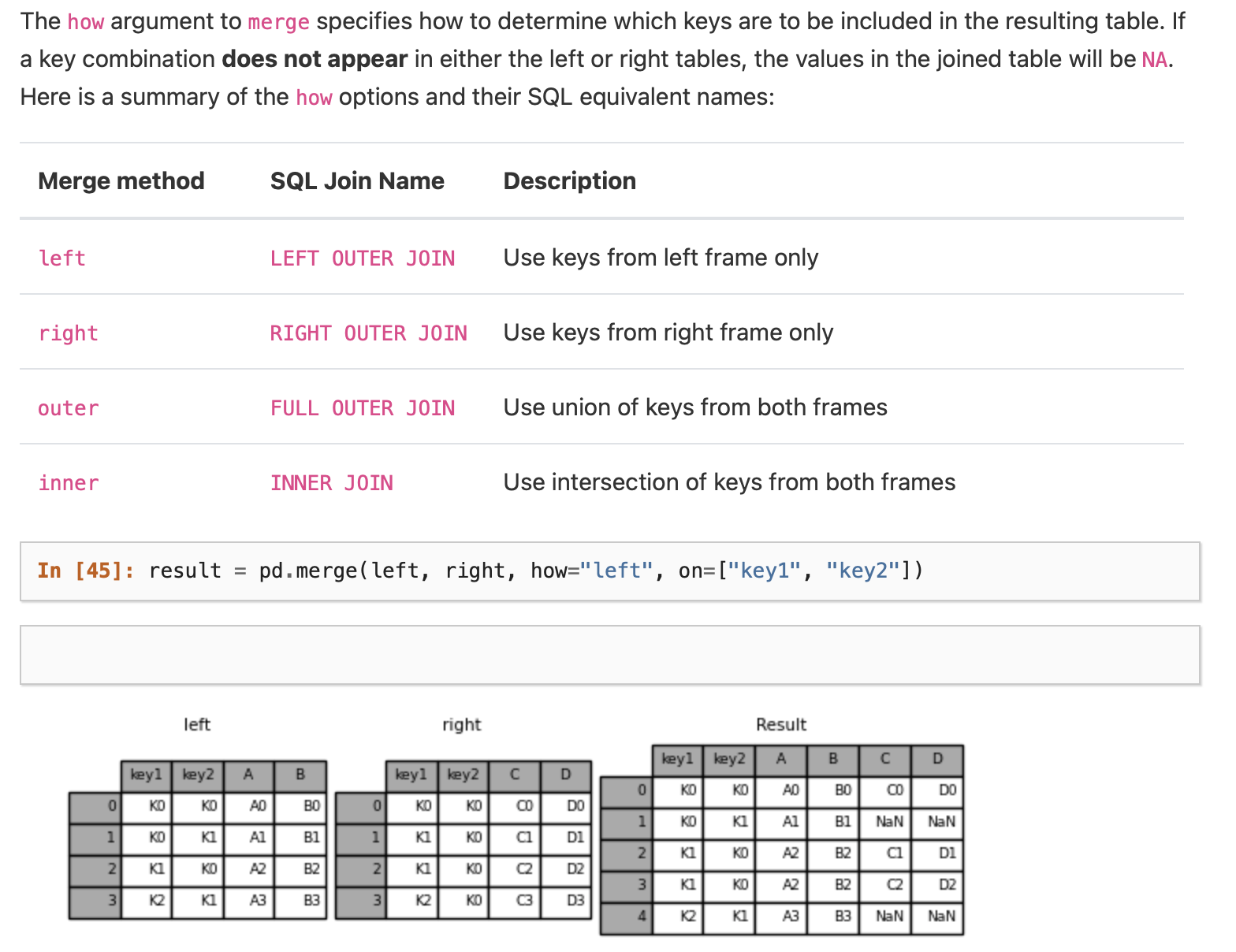
Using .merge()#
User Guide on Merging Dataframes on the .merge() function
Visualizing Data using seaborn and pandas#
This is a preview of content coming later!
# You may need to install seaborn:
# Remember the command to install a package `conda install -c conda-forge seaborn`
import seaborn as sns
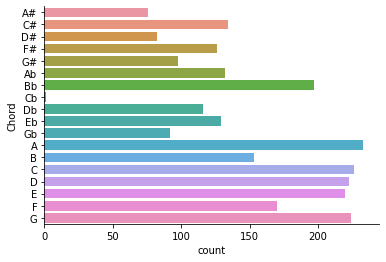
sns.countplot(y="Chord", data=chords)
sns.despine()
Additional Notes on Handling Missing Data#
Attribution#

This notebook contains an excerpt from the Python Data Science Handbook by Jake VanderPlas; the content is available on GitHub.
The text is released under the CC-BY-NC-ND license, and code is released under the MIT license. If you find this content useful, please consider supporting the work by buying the book!
The difference between data found in many tutorials and data in the real world is that real-world data is rarely clean and homogeneous. In particular, many interesting datasets will have some amount of data missing. To make matters even more complicated, different data sources may indicate missing data in different ways.
In this section, we will discuss some general considerations for missing data, discuss how Pandas chooses to represent it, and demonstrate some built-in Pandas tools for handling missing data in Python. Here and throughout the book, we’ll refer to missing data in general as null, NaN, or NA values.
Trade-Offs in Missing Data Conventions#
There are a number of schemes that have been developed to indicate the presence of missing data in a table or DataFrame. Generally, they revolve around one of two strategies: using a mask that globally indicates missing values, or choosing a sentinel value that indicates a missing entry.
In the masking approach, the mask might be an entirely separate Boolean array, or it may involve appropriation of one bit in the data representation to locally indicate the null status of a value.
In the sentinel approach, the sentinel value could be some data-specific convention, such as indicating a missing integer value with -9999 or some rare bit pattern, or it could be a more global convention, such as indicating a missing floating-point value with NaN (Not a Number), a special value which is part of the IEEE floating-point specification.
None of these approaches is without trade-offs: use of a separate mask array requires allocation of an additional Boolean array, which adds overhead in both storage and computation. A sentinel value reduces the range of valid values that can be represented, and may require extra (often non-optimized) logic in CPU and GPU arithmetic. Common special values like NaN are not available for all data types.
As in most cases where no universally optimal choice exists, different languages and systems use different conventions. For example, the R language uses reserved bit patterns within each data type as sentinel values indicating missing data, while the SciDB system uses an extra byte attached to every cell which indicates a NA state.
Missing Data in Pandas#
The way in which Pandas handles missing values is constrained by its reliance on the NumPy package, which does not have a built-in notion of NA values for non-floating-point data types.
Pandas could have followed R’s lead in specifying bit patterns for each individual data type to indicate nullness, but this approach turns out to be rather unwieldy. While R contains four basic data types, NumPy supports far more than this: for example, while R has a single integer type, NumPy supports fourteen basic integer types once you account for available precisions, signedness, and endianness of the encoding. Reserving a specific bit pattern in all available NumPy types would lead to an unwieldy amount of overhead in special-casing various operations for various types, likely even requiring a new fork of the NumPy package. Further, for the smaller data types (such as 8-bit integers), sacrificing a bit to use as a mask will significantly reduce the range of values it can represent.
NumPy does have support for masked arrays – that is, arrays that have a separate Boolean mask array attached for marking data as “good” or “bad.” Pandas could have derived from this, but the overhead in both storage, computation, and code maintenance makes that an unattractive choice.
With these constraints in mind, Pandas chose to use sentinels for missing data, and further chose to use two already-existing Python null values: the special floating-point NaN value, and the Python None object.
This choice has some side effects, as we will see, but in practice ends up being a good compromise in most cases of interest.
None: Pythonic missing data#
The first sentinel value used by Pandas is None, a Python singleton object that is often used for missing data in Python code.
Because it is a Python object, None cannot be used in any arbitrary NumPy/Pandas array, but only in arrays with data type 'object' (i.e., arrays of Python objects):
import numpy as np
import pandas as pd
vals1 = np.array([1, None, 3, 4])
vals1
This dtype=object means that the best common type representation NumPy could infer for the contents of the array is that they are Python objects.
While this kind of object array is useful for some purposes, any operations on the data will be done at the Python level, with much more overhead than the typically fast operations seen for arrays with native types:
for dtype in ["object", "int"]:
print("dtype =", dtype)
%timeit np.arange(1E6, dtype=dtype).sum()
print()
The use of Python objects in an array also means that if you perform aggregations like sum() or min() across an array with a None value, you will generally get an error:
vals1
1 + None
vals1.sum()
This reflects the fact that addition between an integer and None is undefined.
NaN: Missing numerical data#
The other missing data representation, NaN (acronym for Not a Number), is different; it is a special floating-point value recognized by all systems that use the standard IEEE floating-point representation:
vals2 = np.array([1, np.nan, 3, 4])
vals2.dtype
Notice that NumPy chose a native floating-point type for this array: this means that unlike the object array from before, this array supports fast operations pushed into compiled code.
You should be aware that NaN is a bit like a data virus–it infects any other object it touches.
Regardless of the operation, the result of arithmetic with NaN will be another NaN:
1 + np.nan
0 * np.nan
Note that this means that aggregates over the values are well defined (i.e., they don’t result in an error) but not always useful:
vals2.sum(), vals2.min(), vals2.max()
NumPy does provide some special aggregations that will ignore these missing values:
np.nansum(vals2), np.nanmin(vals2), np.nanmax(vals2)
Keep in mind that NaN is specifically a floating-point value; there is no equivalent NaN value for integers, strings, or other types.
NaN and None in Pandas#
NaN and None both have their place, and Pandas is built to handle the two of them nearly interchangeably, converting between them where appropriate:
pd.Series([1, np.nan, 2, None])
For types that don’t have an available sentinel value, Pandas automatically type-casts when NA values are present.
For example, if we set a value in an integer array to np.nan, it will automatically be upcast to a floating-point type to accommodate the NA:
x = pd.Series(range(2), dtype=int)
x
x[0] = None
x
Notice that in addition to casting the integer array to floating point, Pandas automatically converts the None to a NaN value.
(Be aware that there is a proposal to add a native integer NA to Pandas in the future; as of this writing, it has not been included).
While this type of magic may feel a bit hackish compared to the more unified approach to NA values in domain-specific languages like R, the Pandas sentinel/casting approach works quite well in practice and in my experience only rarely causes issues.
The following table lists the upcasting conventions in Pandas when NA values are introduced:
Typeclass |
Conversion When Storing NAs |
NA Sentinel Value |
|---|---|---|
|
No change |
|
|
No change |
|
|
Cast to |
|
|
Cast to |
|
Keep in mind that in Pandas, string data is always stored with an object dtype.
Operating on Null Values#
As we have seen, Pandas treats None and NaN as essentially interchangeable for indicating missing or null values.
To facilitate this convention, there are several useful methods for detecting, removing, and replacing null values in Pandas data structures.
They are:
isnull(): Generate a boolean mask indicating missing valuesnotnull(): Opposite ofisnull()dropna(): Return a filtered version of the datafillna(): Return a copy of the data with missing values filled or imputed
We will conclude this section with a brief exploration and demonstration of these routines.
Detecting null values#
Pandas data structures have two useful methods for detecting null data: isnull() and notnull().
Either one will return a Boolean mask over the data. For example:
data = pd.DataFrame([1, np.nan, "hello", None])
data
data[data.notnull()]
data.isnull()
As mentioned in Data Indexing and Selection, Boolean masks can be used directly as a Series or DataFrame index:
data[data.notnull()]
The isnull() and notnull() methods produce similar Boolean results for DataFrames.
Dropping null values#
In addition to the masking used before, there are the convenience methods, dropna()
(which removes NA values) and fillna() (which fills in NA values). For a Series,
the result is straightforward:
data
data.dropna().reset_index(drop=True)
For a DataFrame, there are more options.
Consider the following DataFrame:
df = pd.DataFrame([[1, np.nan, 2], [2, 3, 5], [np.nan, 4, 6]])
df
We cannot drop single values from a DataFrame; we can only drop full rows or full columns.
Depending on the application, you might want one or the other, so dropna() gives a number of options for a DataFrame.
By default, dropna() will drop all rows in which any null value is present:
df.dropna()
Alternatively, you can drop NA values along a different axis; axis=1 drops all columns containing a null value:
df.dropna(axis="columns")
But this drops some good data as well; you might rather be interested in dropping rows or columns with all NA values, or a majority of NA values.
This can be specified through the how or thresh parameters, which allow fine control of the number of nulls to allow through.
The default is how='any', such that any row or column (depending on the axis keyword) containing a null value will be dropped.
You can also specify how='all', which will only drop rows/columns that are all null values:
df[3] = np.nan
df
df.dropna(axis="columns", how="all")
For finer-grained control, the thresh parameter lets you specify a minimum number of non-null values for the row/column to be kept:
df.dropna(axis="rows", thresh=3)
Here the first and last row have been dropped, because they contain only two non-null values.
Filling null values#
Sometimes rather than dropping NA values, you’d rather replace them with a valid value.
This value might be a single number like zero, or it might be some sort of imputation or interpolation from the good values.
You could do this in-place using the isnull() method as a mask, but because it is such a common operation Pandas provides the fillna() method, which returns a copy of the array with the null values replaced.
Consider the following Series:
data = pd.Series([1, np.nan, 2, None, 3], index=list("abcde"))
data
np.mean(data)
We can fill NA entries with a single value, such as zero:
data.fillna(0)
data.fillna(0).mean()
We can specify a forward-fill to propagate the previous value forward:
# forward-fill
data.fillna(method="ffill")
Or we can specify a back-fill to propagate the next values backward:
# back-fill
data.fillna(method="bfill")
For DataFrames, the options are similar, but we can also specify an axis along which the fills take place:
df
df.fillna(method="ffill", axis="columns")
Notice that if a previous value is not available during a forward fill, the NA value remains.
Combining Datasets: Concat and Append#
Some of the most interesting studies of data come from combining different data sources.
These operations can involve anything from very straightforward concatenation of two different datasets, to more complicated database-style joins and merges that correctly handle any overlaps between the datasets.
Series and DataFrames are built with this type of operation in mind, and Pandas includes functions and methods that make this sort of data wrangling fast and straightforward.
Here we’ll take a look at simple concatenation of Series and DataFrames with the pd.concat function; later we’ll dive into more sophisticated in-memory merges and joins implemented in Pandas.
We begin with the standard imports:
import pandas as pd
import numpy as np
For convenience, we’ll define this function which creates a DataFrame of a particular form that will be useful below:
def make_df(cols, ind):
"""Quickly make a DataFrame"""
data = {c: [str(c) + str(i) for i in ind] for c in cols}
return pd.DataFrame(data, ind)
# example DataFrame
make_df("ABC", range(3))
In addition, we’ll create a quick class that allows us to display multiple DataFrames side by side. The code makes use of the special _repr_html_ method, which IPython uses to implement its rich object display:
class display(object):
"""Display HTML representation of multiple objects"""
template = """<div style="float: left; padding: 10px;">
<p style='font-family:"Courier New", Courier, monospace'>{0}</p>{1}
</div>"""
def __init__(self, *args):
self.args = args
def _repr_html_(self):
return "\n".join(
self.template.format(a, eval(a)._repr_html_()) for a in self.args
)
def __repr__(self):
return "\n\n".join(a + "\n" + repr(eval(a)) for a in self.args)
The use of this will become clearer as we continue our discussion in the following section.
Recall: Concatenation of NumPy Arrays#
Concatenation of Series and DataFrame objects is very similar to concatenation of Numpy arrays, which can be done via the np.concatenate function as discussed in The Basics of NumPy Arrays.
Recall that with it, you can combine the contents of two or more arrays into a single array:
x = [1, 2, 3]
y = [4, 5, 6]
z = [7, 8, 9]
np.concatenate([x, y, z])
The first argument is a list or tuple of arrays to concatenate.
Additionally, it takes an axis keyword that allows you to specify the axis along which the result will be concatenated:
x = [[1, 2], [3, 4]]
np.concatenate([x, x], axis=1)
Simple Concatenation with pd.concat#
Pandas has a function, pd.concat(), which has a similar syntax to np.concatenate but contains a number of options that we’ll discuss momentarily:
# Signature in Pandas v0.18
pd.concat(objs, axis=0, join='outer', join_axes=None, ignore_index=False,
keys=None, levels=None, names=None, verify_integrity=False,
copy=True)
pd.concat() can be used for a simple concatenation of Series or DataFrame objects, just as np.concatenate() can be used for simple concatenations of arrays:
ser1 = pd.Series(["A", "B", "C"], index=[1, 2, 3])
ser2 = pd.Series(["D", "E", "F"], index=[4, 5, 6])
pd.concat([ser1, ser2])
It also works to concatenate higher-dimensional objects, such as DataFrames:
df1 = make_df("AB", [1, 2])
df2 = make_df("AB", [3, 4])
display("df1", "df2", "pd.concat([df1, df2])")
By default, the concatenation takes place row-wise within the DataFrame (i.e., axis='rows').
Like np.concatenate, pd.concat allows specification of an axis along which concatenation will take place.
Consider the following example:
df3 = make_df("AB", [0, 1])
df4 = make_df("CD", [0, 1])
display("df3", "df4", "pd.concat([df3, df4], axis='col')")
We could have equivalently specified axis=1; here we’ve used the more intuitive axis='col'.
Duplicate indices#
One important difference between np.concatenate and pd.concat is that Pandas concatenation preserves indices, even if the result will have duplicate indices!
Consider this simple example:
x = make_df("AB", [0, 1])
y = make_df("AB", [2, 3])
y.index = x.index # make duplicate indices!
display("x", "y", "pd.concat([x, y])")
Notice the repeated indices in the result.
While this is valid within DataFrames, the outcome is often undesirable.
pd.concat() gives us a few ways to handle it.
Catching the repeats as an error#
If you’d like to simply verify that the indices in the result of pd.concat() do not overlap, you can specify the verify_integrity flag.
With this set to True, the concatenation will raise an exception if there are duplicate indices.
Here is an example, where for clarity we’ll catch and print the error message:
pd.concat([x, y])
try:
pd.concat([x, y], verify_integrity=True)
except ValueError as e:
print("ValueError:", e)
Ignoring the index#
Sometimes the index itself does not matter, and you would prefer it to simply be ignored.
This option can be specified using the ignore_index flag.
With this set to true, the concatenation will create a new integer index for the resulting Series:
display("x", "y", "pd.concat([x, y], ignore_index=True)")
Adding MultiIndex keys#
Another option is to use the keys option to specify a label for the data sources; the result will be a hierarchically indexed series containing the data:
display("x", "y", "pd.concat([x, y], keys=['x', 'y'])")
The result is a multiply indexed DataFrame, and we can use the tools discussed in Hierarchical Indexing to transform this data into the representation we’re interested in.
Concatenation with joins#
In the simple examples we just looked at, we were mainly concatenating DataFrames with shared column names.
In practice, data from different sources might have different sets of column names, and pd.concat offers several options in this case.
Consider the concatenation of the following two DataFrames, which have some (but not all!) columns in common:
df5 = make_df("ABC", [1, 2])
df6 = make_df("BCD", [3, 4])
display("df5", "df6", "pd.concat([df5, df6])")
By default, the entries for which no data is available are filled with NA values.
To change this, we can specify one of several options for the join and join_axes parameters of the concatenate function.
By default, the join is a union of the input columns (join='outer'), but we can change this to an intersection of the columns using join='inner':
display("df5", "df6", "pd.concat([df5, df6], join='inner')")
Another option is to directly specify the index of the remaininig colums using the join_axes argument, which takes a list of index objects.
Here we’ll specify that the returned columns should be the same as those of the first input:
display(‘df5’, ‘df6’, “pd.concat([df5, df6], join_axes=[df5.columns])”)
The combination of options of the pd.concat function allows a wide range of possible behaviors when joining two datasets; keep these in mind as you use these tools for your own data.
The append() method#
Because direct array concatenation is so common, Series and DataFrame objects have an append method that can accomplish the same thing in fewer keystrokes.
For example, rather than calling pd.concat([df1, df2]), you can simply call df1.append(df2):
display("df1", "df2", "df1.append(df2)")
Keep in mind that unlike the append() and extend() methods of Python lists, the append() method in Pandas does not modify the original object–instead it creates a new object with the combined data.
It also is not a very efficient method, because it involves creation of a new index and data buffer.
Thus, if you plan to do multiple append operations, it is generally better to build a list of DataFrames and pass them all at once to the concat() function.
In the next section, we’ll look at another more powerful approach to combining data from multiple sources, the database-style merges/joins implemented in pd.merge.
For more information on concat(), append(), and related functionality, see the “Merge, Join, and Concatenate” section of the Pandas documentation.

