Lecture 3 - dates & times, strings, as well as factors
Contents
Lecture 3 - dates & times, strings, as well as factors#
Lecture learning objectives:#
By the end of this lecture and worksheet 3, students should be able to:
Manipulate dates and times using the {lubridate} package
Be able to modify strings in a data frame using regular expressions and the {stringr} package
Cast categorical columns in a data frame as factors when appropriate, and manipulate factor levels as needed in preparation for data visualisation and statistical analysis (using base R and {forcats} package functions)
library(tidyverse)
library(gapminder)
options(repr.matrix.max.rows = 10)
Error in library(tidyverse): there is no package called ‘tidyverse’
Traceback:
1. library(tidyverse)
Tibbles versus data frames#
You have seen and heard me talk about data frames and tibbles in R, and sometimes I carelessly interchange the terms. Let’s take moment to discuss the difference between the two!
Tibbles are special data frames, with special/extra properties. The two important ones you will care about are:
In RStudio, tibbles only output the first 10 rows
When you numerically subset a data frame to 1 column, you get a vector. However, when you numerically subset a tibble you still get a tibble back.
When you create a data frame using base R functions, either via data.frame or one of the base R read.* functions, you get an objects whose class is data frame:
example <- data.frame(a = c(1, 5, 9), b = "z", "a", "t")
example
class(example)
| a | b | X.a. | X.t. |
|---|---|---|---|
| <dbl> | <chr> | <chr> | <chr> |
| 1 | z | a | t |
| 5 | z | a | t |
| 9 | z | a | t |
Tibbles inherit from the data frame class (meaning that have many of the same properties as data frames), but they also have the extra properties I just discussed:
example2 <- tibble(a = c(1, 5, 9), b = "z", "a", "t")
example
class(example2)
| a | b | X.a. | X.t. |
|---|---|---|---|
| <dbl> | <chr> | <chr> | <chr> |
| 1 | z | a | t |
| 5 | z | a | t |
| 9 | z | a | t |
- 'tbl_df'
- 'tbl'
- 'data.frame'
Note: there are some tidyverse functions that will coerce a data frame to a tibble, because what the user is asking for is not possible with a data frame. One such example is group_by (which we will learn about next week):
group_by(example, a) %>%
class()
- 'grouped_df'
- 'tbl_df'
- 'tbl'
- 'data.frame'
Rule of thumb: if you want a tibble, it’s on you to know that and express that explicitly with as_tibble() (if it’s a data frame to start out with).
Dates and times#
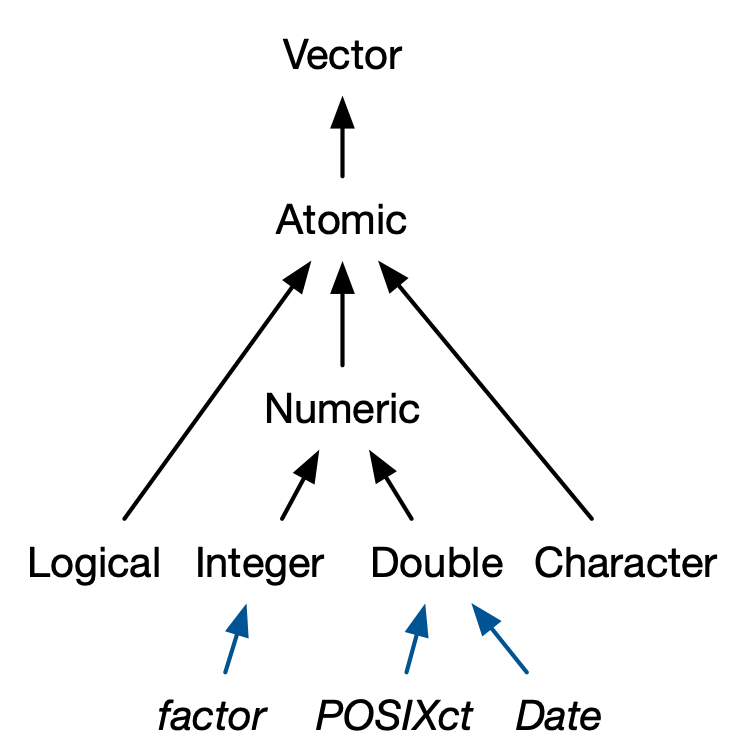
Source: Advanced R by Hadley Wickham
Working with dates and times#
Your weapon: The lubridate package (CRAN; GitHub; main vignette).
library(lubridate)
Attaching package: ‘lubridate’
The following objects are masked from ‘package:base’:
date, intersect, setdiff, union
Get your hands on some dates or date-times#
Use lubridate’s today to get today’s date, without any time:
today()
class(today())
Use lubridate’s now to get RIGHT NOW, meaning the date and the time:
now()
[1] "2022-02-01 09:49:35 PST"
class(now())
- 'POSIXct'
- 'POSIXt'
Get date or date-time from character#
Use the lubridate helpers to convert character or unquoted numbers into dates or date-times:
ymd("2017-01-31")
mdy("January 31st, 2017")
dmy("31-Jan-2017")
ymd(20170131)
ymd_hms("2017-01-31 20:11:59")
[1] "2017-01-31 20:11:59 UTC"
mdy_hm("01/31/2017 08:01")
[1] "2017-01-31 08:01:00 UTC"
You can also force the creation of a date-time from a date by supplying a timezone:
class(ymd(20170131, tz = "UTC"))
- 'POSIXct'
- 'POSIXt'
Build date or date-time from parts#
Instead of a single string, sometimes you’ll have the individual components of the date-time spread across multiple columns.
dates <- tibble(year = c(2015, 2016, 2017, 2018, 2019),
month = c(9, 9, 9, 9, 9),
day = c(3, 4, 2, 6, 3))
dates
| year | month | day |
|---|---|---|
| <dbl> | <dbl> | <dbl> |
| 2015 | 9 | 3 |
| 2016 | 9 | 4 |
| 2017 | 9 | 2 |
| 2018 | 9 | 6 |
| 2019 | 9 | 3 |
To create a date/time from this sort of input, use make_date for dates, or make_datetime for date-times:
# make a single date from year, month and day
dates %>%
mutate(date = make_date(year, month, day))
| year | month | day | date |
|---|---|---|---|
| <dbl> | <dbl> | <dbl> | <date> |
| 2015 | 9 | 3 | 2015-09-03 |
| 2016 | 9 | 4 | 2016-09-04 |
| 2017 | 9 | 2 | 2017-09-02 |
| 2018 | 9 | 6 | 2018-09-06 |
| 2019 | 9 | 3 | 2019-09-03 |
Getting components from a date or date-time#
Sometimes you have the date or date-time and you want to extract a component, such as year or day.
datetime <- ymd_hms("2016-07-08 12:34:56")
datetime
[1] "2016-07-08 12:34:56 UTC"
year(datetime)
month(datetime)
mday(datetime)
yday(datetime)
wday(datetime, label = TRUE, abbr = FALSE)
Levels:
- 'Sunday'
- 'Monday'
- 'Tuesday'
- 'Wednesday'
- 'Thursday'
- 'Friday'
- 'Saturday'
For month and wday you can set label = TRUE to return the abbreviated name of the month or day of the week. Set abbr = FALSE to return the full name.
More date and time wrangling possibilities:#
Tools for working with time spans, to calculate durations, periods and intervals
String manipulations#
The recommended tools:#
stringr package: A core package in the tidyverse. Main functions start with str_. Auto-complete is your friend.
stringrcheatsheettidyr package: Useful for functions that split one character vector into many and vice versa:
separate,unite,extract.tidyrcheatsheetBase functions:
nchar,strsplit,substr,paste, andpaste0.The glue package is fantastic for string interpolation. If stringr::str_interp() doesn’t get your job done, check out the glue package.
Regex-free string manipulation with stringr and tidyr#
Basic string manipulation tasks:
Study a single character vector
How long are the strings?
Presence/absence of a literal string
Operate on a single character vector
Keep/discard elements that contain a literal string
Split into two or more character vectors using a fixed delimiter
Snip out pieces of the strings based on character position
Collapse into a single string
Operate on two or more character vectors
Glue them together element-wise to get a new character vector.
fruit, words, and sentences are character vectors that ship with stringr for practicing.
NOTE - we will be working with vectors today. If you want to operate on data frames, you will need to use these functions inside of data frame/tibble functions, like filter and mutate.
Detect or filter on a target string#
Determine presence/absence of a literal string with str_detect. Spoiler: later we see str_detect also detects regular expressions.
Which fruits actually use the word “fruit”?
# detect "fruit"
#typeof(fruit)
#fruit
str_detect(fruit, "fruit")
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- TRUE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- TRUE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- TRUE
- FALSE
- FALSE
- FALSE
- TRUE
- FALSE
- FALSE
- TRUE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- TRUE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- TRUE
- FALSE
- FALSE
- FALSE
- TRUE
- FALSE
What’s the easiest way to get the actual fruits that match? Use str_subset to keep only the matching elements. Note we are storing this new vector my_fruit to use in later examples!
# subset "fruit"
my_fruit <- str_subset(fruit, "fruit")
my_fruit
- 'breadfruit'
- 'dragonfruit'
- 'grapefruit'
- 'jackfruit'
- 'kiwi fruit'
- 'passionfruit'
- 'star fruit'
- 'ugli fruit'
String splitting by delimiter#
Use stringr::str_split to split strings on a delimiter.
Some of our fruits are compound words, like “grapefruit”, but some have two words, like “ugli fruit”. Here we split on a single space ” “, but show use of a regular expression later.
# split on " "
str_split(my_fruit, " ")
- 'breadfruit'
- 'dragonfruit'
- 'grapefruit'
- 'jackfruit'
-
- 'kiwi'
- 'fruit'
- 'passionfruit'
-
- 'star'
- 'fruit'
-
- 'ugli'
- 'fruit'
It’s bummer that we get a list back. But it must be so! In full generality, split strings must return list, because who knows how many pieces there will be?
If you are willing to commit to the number of pieces, you can use str_split_fixed and get a character matrix. You’re welcome!
str_split_fixed(my_fruit, pattern = " ", n = 2)
| breadfruit | |
| dragonfruit | |
| grapefruit | |
| jackfruit | |
| kiwi | fruit |
| passionfruit | |
| star | fruit |
| ugli | fruit |
If the to-be-split variable lives in a data frame, tidyr::separate will split it into 2 or more variables:
# separate on " "
my_fruit[5] <- "yellow kiwi fruit"
my_fruit
tibble(unsplit = my_fruit) %>%
separate(unsplit, into = c("pre", "post"), sep = " ")
- 'breadfruit'
- 'dragonfruit'
- 'grapefruit'
- 'jackfruit'
- 'yellow kiwi fruit'
- 'passionfruit'
- 'star fruit'
- 'ugli fruit'
Warning message:
“Expected 2 pieces. Additional pieces discarded in 1 rows [5].”
Warning message:
“Expected 2 pieces. Missing pieces filled with `NA` in 5 rows [1, 2, 3, 4, 6].”
| pre | post |
|---|---|
| <chr> | <chr> |
| breadfruit | NA |
| dragonfruit | NA |
| grapefruit | NA |
| jackfruit | NA |
| yellow | kiwi |
| passionfruit | NA |
| star | fruit |
| ugli | fruit |
Substring extraction (and replacement) by position#
Count characters in your strings with str_length. Note this is different from the length of the character vector itself.
# get length of each string
str_length(my_fruit)
my_fruit
- 10
- 11
- 10
- 9
- 17
- 12
- 10
- 10
- 'breadfruit'
- 'dragonfruit'
- 'grapefruit'
- 'jackfruit'
- 'yellow kiwi fruit'
- 'passionfruit'
- 'star fruit'
- 'ugli fruit'
You can snip out substrings based on character position with str_sub.
# remove first three strings
str_sub(my_fruit, 1, 3)
- 'bre'
- 'dra'
- 'gra'
- 'jac'
- 'yel'
- 'pas'
- 'sta'
- 'ugl'
Finally, str_sub also works for assignment, i.e. on the left hand side of <-
# replace three characters with AAA
str_sub(my_fruit, 1, 3) <- "AAA"
my_fruit
- 'AAAadfruit'
- 'AAAgonfruit'
- 'AAApefruit'
- 'AAAkfruit'
- 'AAAlow kiwi fruit'
- 'AAAsionfruit'
- 'AAAr fruit'
- 'AAAi fruit'
Collapse a vector#
You can collapse a character vector of length n > 1 to a single string with str_c, which also has other uses (see the next section).
# collapse a character vector into one
head(fruit) %>%
str_c(collapse = "-")
Create a character vector by catenating multiple vectors#
If you have two or more character vectors of the same length, you can glue them together element-wise, to get a new vector of that length. Here are some … awful smoothie flavors?
# concatenate character vectors
fruit[1:4]
fruit[5:8]
str_c(fruit[1:4], fruit[5:8], sep = "")
- 'apple'
- 'apricot'
- 'avocado'
- 'banana'
- 'bell pepper'
- 'bilberry'
- 'blackberry'
- 'blackcurrant'
- 'applebell pepper'
- 'apricotbilberry'
- 'avocadoblackberry'
- 'bananablackcurrant'
If the to-be-combined vectors are variables in a data frame, you can use tidyr::unite to make a single new variable from them.
# concatenate character vectors when they are in a data frame
tibble(fruit1 = fruit[1:4],
fruit2 = fruit[5:8]) %>%
unite("flavour_combo", fruit1, fruit2, sep = " & ")
| flavour_combo |
|---|
| <chr> |
| apple & bell pepper |
| apricot & bilberry |
| avocado & blackberry |
| banana & blackcurrant |
Substring replacement#
You can replace a pattern with str_replace. Here we use an explicit string-to-replace, but later we revisit with a regular expression.
# replace fruit with vegetable
my_fruit <- str_subset(fruit, "fruit")
my_fruit
str_replace(my_fruit, "fruit", "vegetable")
- 'breadfruit'
- 'dragonfruit'
- 'grapefruit'
- 'jackfruit'
- 'kiwi fruit'
- 'passionfruit'
- 'star fruit'
- 'ugli fruit'
- 'breadvegetable'
- 'dragonvegetable'
- 'grapevegetable'
- 'jackvegetable'
- 'kiwi vegetable'
- 'passionvegetable'
- 'star vegetable'
- 'ugli vegetable'
A special case that comes up a lot is replacing
NA, for which there isstr_replace_na.If the NA-afflicted variable lives in a data frame, you can use
tidyr::replace_na.
Other str_* functions?#
There are many many other useful str_* functions from the stringr package. Too many to go through them all here. If these shown in lecture aren’t what you need, then you should try ?str + tab to see the possibilities:
?str_
Regular expressions with stringr#
or…

Examples with gapminder#
library(gapminder)
head(gapminder)
| country | continent | year | lifeExp | pop | gdpPercap |
|---|---|---|---|---|---|
| <fct> | <fct> | <int> | <dbl> | <int> | <dbl> |
| Afghanistan | Asia | 1952 | 28.801 | 8425333 | 779.4453 |
| Afghanistan | Asia | 1957 | 30.332 | 9240934 | 820.8530 |
| Afghanistan | Asia | 1962 | 31.997 | 10267083 | 853.1007 |
| Afghanistan | Asia | 1967 | 34.020 | 11537966 | 836.1971 |
| Afghanistan | Asia | 1972 | 36.088 | 13079460 | 739.9811 |
| Afghanistan | Asia | 1977 | 38.438 | 14880372 | 786.1134 |
Filtering rows with str_detect#
Let’s filter for rows where the country name starts with “AL”:
library(tidyverse)
library(gapminder)
# detect countries that start with "AL"
gapminder %>%
filter(str_detect(country, "^Al")) %>%
pull(country) %>%
unique() %>%
length()
And now rows where the country ends in tan:
# detect countries that end with "tan"
gapminder %>%
filter(str_detect(country, "tan$"))
| country | continent | year | lifeExp | pop | gdpPercap |
|---|---|---|---|---|---|
| <fct> | <fct> | <int> | <dbl> | <int> | <dbl> |
| Afghanistan | Asia | 1952 | 28.801 | 8425333 | 779.4453 |
| Afghanistan | Asia | 1957 | 30.332 | 9240934 | 820.8530 |
| Afghanistan | Asia | 1962 | 31.997 | 10267083 | 853.1007 |
| Afghanistan | Asia | 1967 | 34.020 | 11537966 | 836.1971 |
| Afghanistan | Asia | 1972 | 36.088 | 13079460 | 739.9811 |
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ |
| Pakistan | Asia | 1987 | 58.245 | 105186881 | 1704.687 |
| Pakistan | Asia | 1992 | 60.838 | 120065004 | 1971.829 |
| Pakistan | Asia | 1997 | 61.818 | 135564834 | 2049.351 |
| Pakistan | Asia | 2002 | 63.610 | 153403524 | 2092.712 |
| Pakistan | Asia | 2007 | 65.483 | 169270617 | 2605.948 |
Or countries containing “, Dem. Rep.” :
# detect countries that contain ", Dem. Rep."
gapminder %>%
filter(str_detect(country, "\\, Dem. Rep."))
| country | continent | year | lifeExp | pop | gdpPercap |
|---|---|---|---|---|---|
| <fct> | <fct> | <int> | <dbl> | <int> | <dbl> |
| Congo, Dem. Rep. | Africa | 1952 | 39.143 | 14100005 | 780.5423 |
| Congo, Dem. Rep. | Africa | 1957 | 40.652 | 15577932 | 905.8602 |
| Congo, Dem. Rep. | Africa | 1962 | 42.122 | 17486434 | 896.3146 |
| Congo, Dem. Rep. | Africa | 1967 | 44.056 | 19941073 | 861.5932 |
| Congo, Dem. Rep. | Africa | 1972 | 45.989 | 23007669 | 904.8961 |
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ |
| Korea, Dem. Rep. | Asia | 1987 | 70.647 | 19067554 | 4106.492 |
| Korea, Dem. Rep. | Asia | 1992 | 69.978 | 20711375 | 3726.064 |
| Korea, Dem. Rep. | Asia | 1997 | 67.727 | 21585105 | 1690.757 |
| Korea, Dem. Rep. | Asia | 2002 | 66.662 | 22215365 | 1646.758 |
| Korea, Dem. Rep. | Asia | 2007 | 67.297 | 23301725 | 1593.065 |
Replace “, Dem. Rep.” with ” Democratic Republic”:
# replace ", Dem. Rep." with " Democratic Republic"
gapminder %>%
mutate(country = str_replace(country,
"\\, Dem. Rep.",
" Democratic Republic")) %>%
filter(country == "Korea Democratic Republic")
| country | continent | year | lifeExp | pop | gdpPercap |
|---|---|---|---|---|---|
| <chr> | <fct> | <int> | <dbl> | <int> | <dbl> |
| Korea Democratic Republic | Asia | 1952 | 50.056 | 8865488 | 1088.278 |
| Korea Democratic Republic | Asia | 1957 | 54.081 | 9411381 | 1571.135 |
| Korea Democratic Republic | Asia | 1962 | 56.656 | 10917494 | 1621.694 |
| Korea Democratic Republic | Asia | 1967 | 59.942 | 12617009 | 2143.541 |
| Korea Democratic Republic | Asia | 1972 | 63.983 | 14781241 | 3701.622 |
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ |
| Korea Democratic Republic | Asia | 1987 | 70.647 | 19067554 | 4106.492 |
| Korea Democratic Republic | Asia | 1992 | 69.978 | 20711375 | 3726.064 |
| Korea Democratic Republic | Asia | 1997 | 67.727 | 21585105 | 1690.757 |
| Korea Democratic Republic | Asia | 2002 | 66.662 | 22215365 | 1646.758 |
| Korea Democratic Republic | Asia | 2007 | 67.297 | 23301725 | 1593.065 |
Extract matches#
To extract the actual text of a match, use str_extract.
We’re going to need a more complicated example, the Harvard sentences from the stringr package:
typeof(sentences)
head(sentences)
- 'The birch canoe slid on the smooth planks.'
- 'Glue the sheet to the dark blue background.'
- 'It\'s easy to tell the depth of a well.'
- 'These days a chicken leg is a rare dish.'
- 'Rice is often served in round bowls.'
- 'The juice of lemons makes fine punch.'
Say we want to extract all of the colours used in the sentences. We can do this by creating a pattern which would match them, and passing that and our vector of sentences to str_extract:
colours <- "red|orange|yellow|green|blue|purple"
# extract colours used in sentences
str_extract(sentences, colours)
- NA
- 'blue'
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- 'blue'
- NA
- 'red'
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- 'red'
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- 'red'
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- 'blue'
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- 'yellow'
- NA
- NA
- NA
- 'red'
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- 'red'
- NA
- 'green'
- 'red'
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- 'red'
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- 'blue'
- 'red'
- NA
- 'red'
- 'red'
- NA
- NA
- NA
- NA
- NA
- 'red'
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- ⋯
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- 'red'
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- 'red'
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- 'green'
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- 'green'
- 'green'
- NA
- NA
- NA
- NA
- NA
- 'red'
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- 'red'
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- 'yellow'
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- 'red'
- NA
- 'orange'
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- 'red'
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- 'red'
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- 'red'
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
- NA
str_extract only returns the first match for each element. To return all matches from an element we need to use str_extract_all:
# extract all colours used in sentences
str_extract_all(sentences, colours)
- 'blue'
- 'blue'
- 'red'
- 'red'
- 'red'
- 'blue'
- 'yellow'
- 'red'
- 'red'
- 'green'
- 'red'
- 'red'
- 'blue'
- 'red'
- 'red'
- 'red'
- 'red'
- 'blue'
- 'red'
-
- 'blue'
- 'red'
- 'red'
- 'green'
- 'red'
- 'red'
- 'red'
- 'red'
- 'red'
- 'red'
- 'green'
- 'red'
- 'green'
- 'red'
- 'purple'
- 'green'
- 'red'
- 'red'
- 'red'
- 'red'
- 'red'
- 'blue'
- 'red'
- 'blue'
- 'red'
- 'red'
- 'red'
- 'red'
- 'green'
- 'green'
-
- 'green'
- 'red'
- 'red'
- 'red'
- 'yellow'
- 'red'
-
- 'orange'
- 'red'
- 'red'
- 'red'
- 'red'
Note: str_extract returns a character vector, whereas str_extract_all returns a litst. This is because when asking for multiple matches back, you do not know how many you will get, and thus we cannot expect a rectangular shape.
Capture groups#
You can also use parentheses to extract parts of a complex match.
For example, imagine we want to extract nouns from the sentences. As a heuristic, we’ll look for any word that comes after “a” or “the”. Defining a “word” in a regular expression is a little tricky, so here I use a simple approximation: a sequence of at least one character that isn’t a space.
# extract nouns from sentences
noun <- "(a|the) ([^ ]+)"
str_match(sentences, noun) %>%
head()
| the smooth | the | smooth |
| the sheet | the | sheet |
| the depth | the | depth |
| a chicken | a | chicken |
| NA | NA | NA |
| NA | NA | NA |
Like str_extract, if you want all matches for each string, you’ll need str_match_all
Summary of string manipulation functions covered so far:#
function |
description |
|---|---|
|
Detects elements in a vector that match a pattern, returns a vector of logicals |
|
Detects and returns elements in a vector that match a pattern |
|
Split strings in a vector on a delimiter. Returns a list (used |
|
Split character vectors from a data frame on a delimiter which get returned as additional columns in the data frame |
|
Counts the number of characters for each element of a character vector, and returns a numeric vector of the counts |
|
Remove substrings based on character position |
|
Collapse and/or concatenate elements from a character vector(s) |
|
Concatenate elements from character vectors from a data frame to create a single column |
|
Replace a pattern in a vector of character vectors with a given string |
|
Extract the actual text of a match from a character vector |
|
Use capture groups to extract parts of a complex match from a character vector, returns the match and the capture groups as columns of a matrix |
Factors#

Source: Advanced R by Hadley Wickham
Be the boss of your factors#
I love and hate factors
I love them for data visualization and statistics because I do not need to make dummy variables
I hate them because if you are not careful, they fool you because they look like character vectors. And when you treat them like character vectors you get cryptic error messages, like we saw when we tried to do a conditional mutate on the gapminder data set
Tidyverse philosophy for factors#
Humans, not computers should decide which columns are factors
Factors are not that useful until you are at the end of your data wrangling, before that you want character vectors so you can do string manipulations
Tidyverse functions, like
tibble, andread_csvgive you columns with strings as character vectors, Base R functions likedata.frameandread.csv
Factor inspection#
Get to know your factor before you start touching it! It’s polite. Let’s use gapminder$continent as our example.
str(gapminder$continent)
Factor w/ 5 levels "Africa","Americas",..: 3 3 3 3 3 3 3 3 3 3 ...
levels(gapminder$continent)
- 'Africa'
- 'Americas'
- 'Asia'
- 'Europe'
- 'Oceania'
nlevels(gapminder$continent)
class(gapminder$continent)
Dropping unused levels#
Just because you drop all the rows corresponding to a specific factor level, the levels of the factor itself do not change. Sometimes all these unused levels can come back to haunt you later, e.g., in figure legends.
Watch what happens to the levels of country when we filter Gapminder to a handful of countries:
nlevels(gapminder$country)
h_countries <- gapminder %>%
filter(country %in% c("Egypt", "Haiti", "Romania", "Thailand", "Venezuela"))
nlevels(h_countries$country)
huh? Even though h_gap only has data for a handful of countries, we are still schlepping around all 142 levels from the original gapminder tibble.
How to get rid of them? We’ll use the forcats::fct_drop function to do this:
h_countries$country %>% nlevels()
h_countries$country %>%
fct_drop() %>%
nlevels
Change order of the levels, principled#
By default, factor levels are ordered alphabetically. Which might as well be random, when you think about it! It is preferable to order the levels according to some principle:
Frequency. Make the most common level the first and so on.
Another variable. Order factor levels according to a summary statistic for another variable. Example: order Gapminder countries by life expectancy.
First, let’s order continent by frequency, forwards and backwards. This is often a great idea for tables and figures, esp. frequency barplots.
## default order is alphabetical
gapminder$continent %>%
levels()
- 'Africa'
- 'Americas'
- 'Asia'
- 'Europe'
- 'Oceania'
Let’s use forcats::fct_infreq to order by frequency:
gapminder$continent %>%
fct_infreq() %>%
levels()
gap2 <- gapminder %>%
mutate(continent = fct_infreq(continent))
gap2$continent %>%
levels()
- 'Africa'
- 'Asia'
- 'Europe'
- 'Americas'
- 'Oceania'
- 'Africa'
- 'Asia'
- 'Europe'
- 'Americas'
- 'Oceania'
Or reverse frequency:
gapminder$continent %>%
fct_infreq() %>%
fct_rev() %>%
levels()
- 'Oceania'
- 'Americas'
- 'Europe'
- 'Asia'
- 'Africa'
Order one variable by another#
You can use forcats::fct_reorder to order one variable by another.
The factor is the grouping variable and the default summarizing function is median but you can specify something else.
## order countries by median life expectancy
fct_reorder(gapminder$country, gapminder$lifeExp) %>%
levels() %>%
head()
- 'Sierra Leone'
- 'Guinea-Bissau'
- 'Afghanistan'
- 'Angola'
- 'Somalia'
- 'Guinea'
Using min instead to reorder the factors:
## order accoring to minimum life exp instead of median
fct_reorder(gapminder$country, gapminder$lifeExp, min) %>%
levels() %>%
head()
- 'Rwanda'
- 'Afghanistan'
- 'Gambia'
- 'Angola'
- 'Sierra Leone'
- 'Cambodia'
Change order of the levels, “because I said so”#
Sometimes you just want to hoist one or more levels to the front. Why? Because I said so (sometimes really useful when creating visualizations).
Reminding ourselves of the level order for gapminder$continent:
gapminder$continent %>% levels()
- 'Africa'
- 'Americas'
- 'Asia'
- 'Europe'
- 'Oceania'
Reorder and put Asia and Africa first:
gapminder$continent %>%
fct_relevel("Asia", "Africa") %>%
levels()
- 'Asia'
- 'Africa'
- 'Americas'
- 'Europe'
- 'Oceania'
Why do we need to know how to do this?#
Factor levels impact statistical analysis & data visualization!
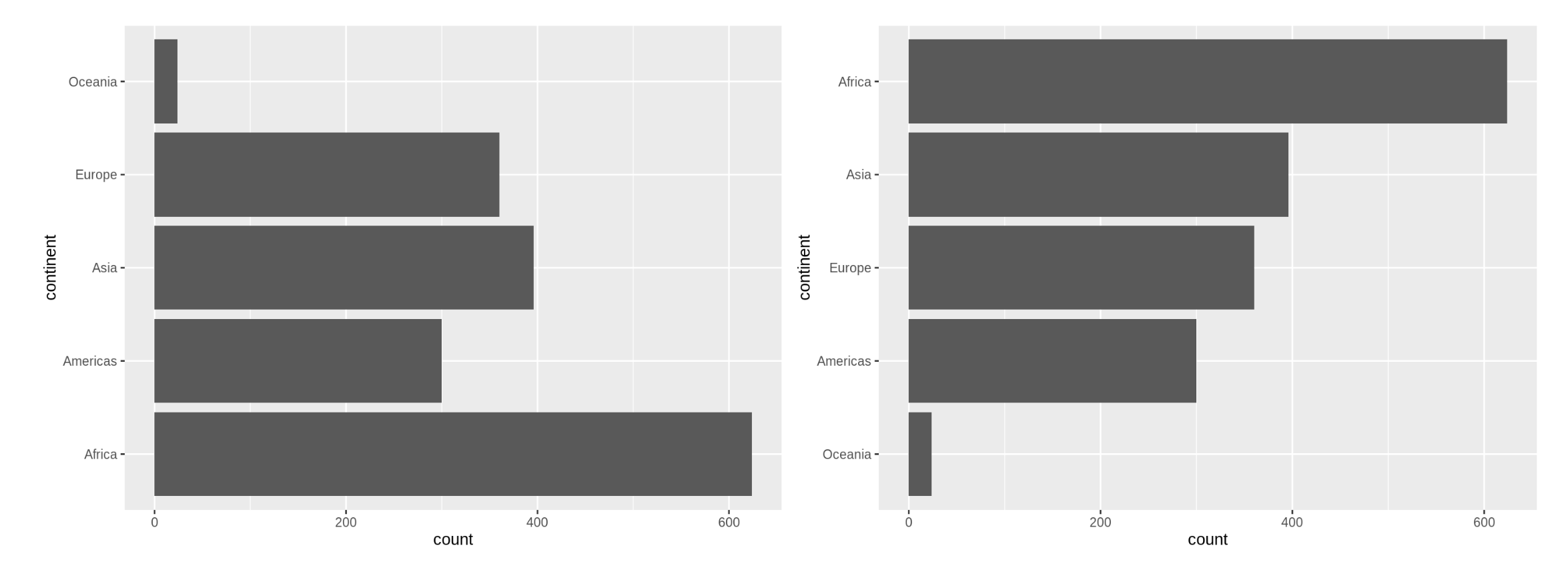
For example, these two barcharts of frequency by continent differ only in the order of the continents. Which do you prefer? Discuss with your neighbour.
What did we learn today?#
The differences between data frames and tibbles
The beautiful {lubridate} package for working with dates and times
Tools for manipulating and working with character data in the {stringr} and {tidyr} packages
How to take control of our factors using {forcats} and how to investigate factors using base R functions
Attributions#
Stat 545 created by Jenny Bryan
R for Data Science by Garrett Grolemund & Hadley Wickham

