Lecture 7: Functional-style programming in R
Contents
Lecture 7: Functional-style programming in R#
Note from Firas
Our series of R lectures will be presented by Dr. Tiffany Timbers, the other option co-director of the Vancouver MDS program.
First, some things leftover from last week…#
Reading in functions from an R script#
Usually the step before packaging your code, is having some functions in another script that you want to read into your analysis. We use the source function to do this:
source("src/kelvin_to_celsius.R")
Warning message in file(filename, "r", encoding = encoding):
“cannot open file 'src/kelvin_to_celsius.R': No such file or directory”
Error in file(filename, "r", encoding = encoding): cannot open the connection
Traceback:
1. source("src/kelvin_to_celsius.R")
2. file(filename, "r", encoding = encoding)
Once you do this, you have access to all functions contained within that script:
kelvin_to_celsius(273.15)
Note - this is how the test_* functions are brought into your Jupyter notebooks for the autograding part of your lab3 homework.
Introduction to R packages#
source("script_with_functions.R")is useful, but when you start using these functions in different projects you need to keep copying the script, or having overly specific paths…
The next step is packaging your R code so that it can be installed and then used across multiple projects on your (and others) machines without directly pointing to where the code is stored, but instead accessed using the
libraryfunction.
You will learn how to do this in Collaborative Software Development (term 2), but for now, let’s tour a simple R package to get a better understanding of what they are: https://github.com/ttimbers/convertemp
Install the convertemp R package:#
In RStudio, type: devtools::install_github("ttimbers/convertemp")
library(convertemp)
?celsius_to_kelvin
celsius_to_kelvin(0)
Packages and environments#
Each package attached by library() becomes one of the parents of the global environment
The immediate parent of the global environment is the last package you attached, the parent of that package is the second to last package you attached, …
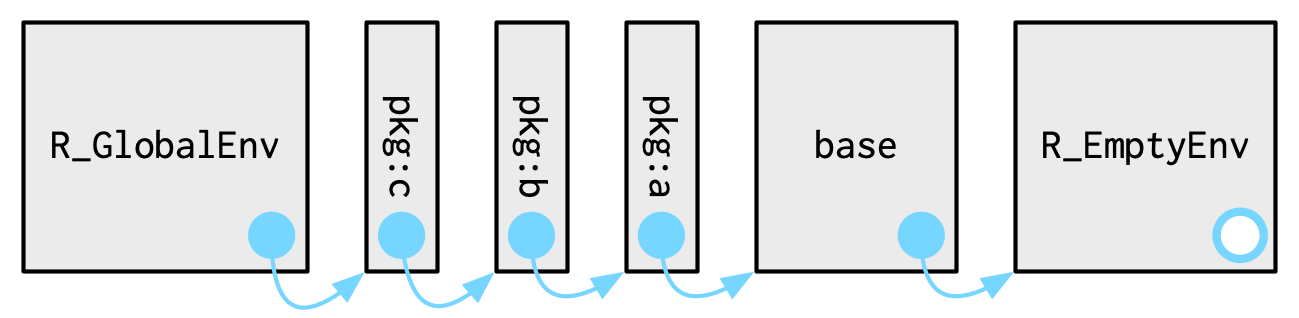
Source: Advanced R by Hadley Wickham
Packages and environments#
When you attach another package with library(), the parent environment of the global environment changes:
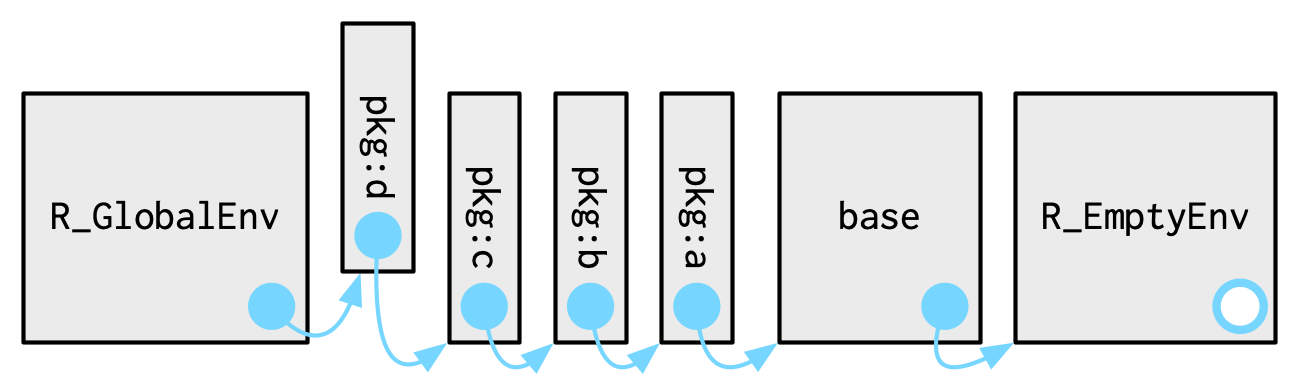
Source: Advanced R by Hadley Wickham
Functional style programming in R with purrr#

If you have programmed in R before#
purrr is an alternative to “apply” functions
purrr::map() ≈ base::lapply()
How do we apply a function to all columns of a data frame?#
Say, for example we wanted to calculate the median for each column in the mtcars data frame:
head(mtcars)
medians <- vector("double", ncol(mtcars))
for (i in seq_along(mtcars)) {
medians[i] <- median(mtcars[[i]], na.rm = TRUE)
}
OK, then next we want to calculate the mean for all of the columns:
means <- vector("double", ncol(mtcars))
for (i in seq_along(mtcars)) {
means[i] <- mean(mtcars[[i]], na.rm = TRUE)
}
OK, and then the variance…
variances <- vector("double", ncol(mtcars))
for (i in seq_along(mtcars)) {
variances[i] <- var(mtcars[[i]], na.rm = TRUE)
}
This is getting a little repetitive… What are we repeating?
Can we write this as a function?#
Given that functions are objects in R, this seems reasonable!
medians <- vector("double", ncol(mtcars))
for (i in seq_along(mtcars)) {
medians[i] <- median(mtcars[[i]], na.rm = TRUE)
}
This is essentially the guts of purrr::map_dbl. The only difference is that is coded in C and the use of ... for additional arguments.
mds_map <- function(x, fun) {
out <- vector("double", ncol(x))
for (i in seq_along(x)) {
out[i] <- fun(x[[i]], na.rm = TRUE)
}
out
}
mds_map(mtcars, min)
Functionals#
We have just written what is called a functional.
A functional is a function that takes a function (and other things) as an input and returns a vector as output.
R has several other functionals outside of purrr that you might have already encountered: lapply, apply, tapply, integrate or optim.
What can you do with functionals?#
Common use is as an alternative to for loops
For loops are actually quite effective for iteration, and efficient when used, however it is easy to make mistakes when setting them up as you have to:
pre-allocate space for the output
iterate over the thing the right amount of times
properly use the iteration index
Of course someone has to write for loops#
It doesn’t have to be you#
– Jenny Bryan, Software Developer at RStudio and MDS Founder
The purrr::map* family of functions#

Source: Advanced R by Hadley Wickham
Let’s start at the beginning with the most general purrr function: map#
map(.x, .f, ...)
Above reads as: for every element of .x apply .f
and can be pictured as:
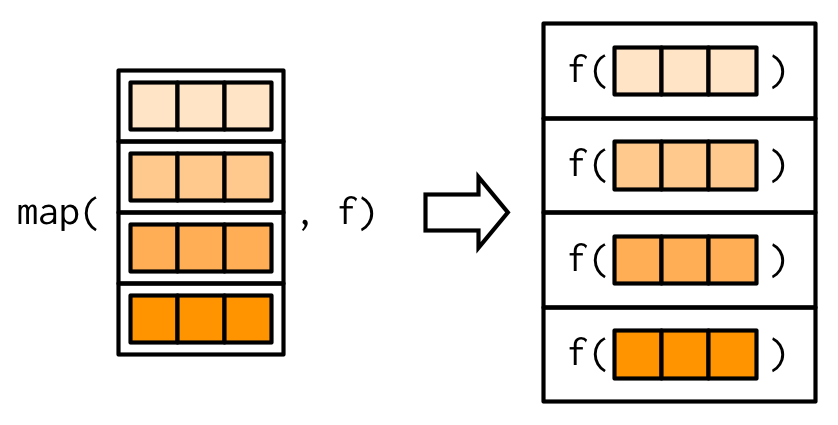
Or picture as…
Source: Row-oriented workflows in R with the tidyverse by Jenny Bryan

Source: Row-oriented workflows in R with the tidyverse by Jenny Bryan
purrr::map test drive#
Let’s calculate the median of all the columns of the mtcars data frame using purrr::map:
library(purrr)
map(mtcars, median)
That looks different from our mds_map function! The output is of type list.
Choosing the purrr::map* function based on your desired output#

Source: Advanced R by Hadley Wickham
Trying again with purrr::map_dbl#
map_dbl(mtcars, median)
What if our data frame had missing values?#
Let’s make some to see the consequences…
mtcars_NA <- mtcars
mtcars_NA[1, 1] <- NA
map_dbl(mtcars_NA, median)
map_dbl returns a vector of type double.
How do we tell median to ignore NA’s? Using na.rm = TRUE! But how do we add this to our map_dbl call?
Solution!#
Creating an anonymous function within the purrr::map_dbl function!
map_dbl(mtcars_NA, function(df) median(df, na.rm = TRUE))
(function(x) x + 1)(1)
Above the function takes in x as an argument and adds one to it. The function definition is surrounded by round brackets, as is the value being passed to the anonymous function.
Aside: Anonymous functions in R#
General format: function(x) body_of_function
To use one in the global environment, outside of another function call, you do the following:
Back to anonymous function calls within purrr::map*#
Long form:
map_dbl(mtcars_NA, function(df) median(df, na.rm = TRUE))
Short form:
map_dbl(mtcars_NA, ~ median(., na.rm = TRUE))
In the shortcut we replace function(VARIABLE) with a ~ and replace the VARIABLE in the function call with a .
Challenge 1:#
Use a purrr::map function to caclulate the variance (using var) of each of the numerical columns in the iris dataset. Return the object as a data frame.
Mapping with > 1 data objects#
What if the function you want to map takes in > 1 data objects?
map2* and pmap* are your friends here!
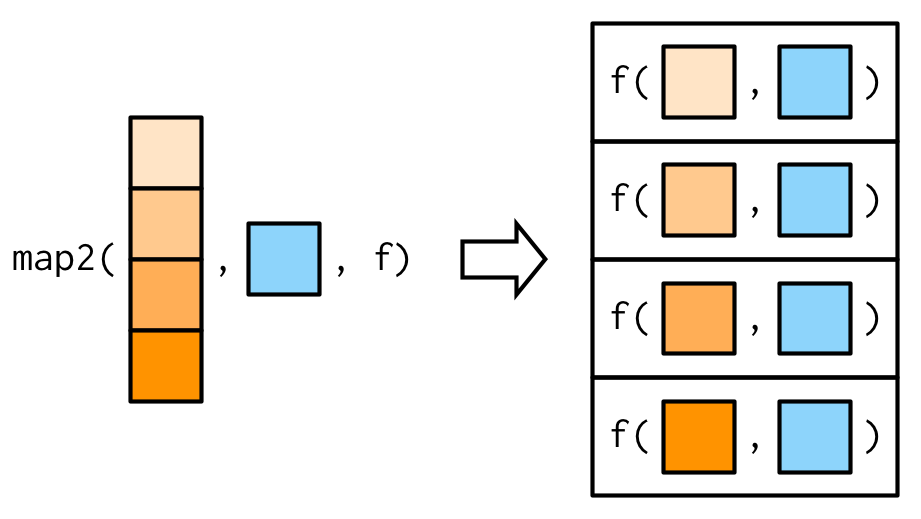
purrr::map2*#
map2*(.x, .y, .f, ...)
Above reads as: for every element of .x and .y apply .f
Or picture as…
Source: purrr workshop by Jenny Bryan

Source: purrr workshop by Jenny Bryan
purrr::map2_df example:#
For example, say you want to calculate a weighted means (using weighted.mean) for columns of a data frame where you had another data frame containing those weights.
Let’s make some data:
data <- tibble(frequency = runif(10),
loudness = runif(10),
power = runif(10),
rating = rpois(10, 5) + 1,
year = rpois(10, 5) + 1999)
data[1, 1] <- NA
data
library(dplyr, quietly = TRUE)
data <- tibble(x1 = runif(10),
x2 = runif(10),
x3 = runif(10))
data[1, 1] <- NA
weights <- tibble(x1 = rpois(10, 5) + 1,
x2 = rpois(10, 5) + 1,
x3 = rpois(10, 5) + 1,)
data
weights
purrr::map2_df example:#
Let’s use map2_df to calculate the weighted mean using these two data frames.
?weighted.mean
map2_df(data, weights, weighted.mean)
Ah! That NA got us again! We need to write this an an anonymous function so that we can pass in na.rm = TRUE
purrr::map2_df example:#
Now using an anonymous function with the long form:
map2_df(data, weights, function(x, y) weighted.mean(x, y, na.rm = TRUE))
Now with the short form:
map2_df(data, weights, ~ weighted.mean(.x, .y, na.rm = TRUE))
Not too bad eh!
purrr::map2*#
Also, if y has less elements than x, it recycles y:
This is most useful when y has only one element.
purrr::pmap*#
pmap*(list(.x1, .x2, ... .xn), .f, ...)
Above reads as: for every element of in the list (that contains .x1, .x2, ... .xn) apply .f
Example of using pmap_df to calculate the weighted means:#
pmap_df(list(data, weights), ~ weighted.mean(.x, .y, na.rm = TRUE))
But what happens when you have > 2 arguments?
More than two arguments#
Without an anonymous function, works as so:
f1 <- function(x, y, z) {
x + y + z
}
pmap_dbl(list(c(1, 1), c(1, 2), c(2, 2)), f1)
If you want to use an anonymous function, then use ..1, ..2, ..3, and so on to specify where the mapped objets go in your function:
f2 <- function(x, y, z, a = 0) {
x + y + z + a
}
pmap_dbl(list(c(1, 1), c(1, 2), c(2, 2)), ~ f2(..1, ..2, ..3, a = -1))
We only used two inputs to our function here, but we can use any number with pmap, we just need to add them to our list!
Want to iterate row-wise, instead of column-wise?#
Here you can use purrr::pmap on a single data frame!
This: purrr::pmap(df, .f)
reads as: for every tuple in .l (i.e., each row of df) apply .f
The key point is that pmap() iterates over tuples = the collection of i-th elements of k lists. A data frame row is an interesting special case.
Here’s an example of row-wise iteration#
Here we calculate the sum for each row in the mtcars data frame:
pmap(mtcars, sum)
What about mapping over groups of rows???#
There are two strategies we will learn in the Data Wrangling course next block:
dplyr::group_by+dplyr::summarizedplyr::group_by+tidyr::nest
What did we learn today?#
Attribution#
Advanced R by Hadley Wickham

